2,561 genes that confer collective resistance to some of the most widely used antibiotics are detected
The global health risk posed by antibiotic resistance genes (ARG) is a challenge for the health system and, in turn, an alert for people. The threat is that the human body does not correctly process information from clinical treatments administered against pathogens. This is a growing reality, which makes the treatment of human diseases increasingly complex, although it also has an effect on animals. The recent study Assessment of global health risk of antibiotic resistance genes, published in Nature Communications, detects 2,561 genes that collectively confer resistance to 24 classes of antibiotics (based on the Comprehensive Antibiotic Research Database) and reaches a conclusion to take into account: 23.78% of these genes pose a risk to human health because they confer resistance to multiple drugs and, as a consequence, increasingly sophisticated clinical treatments. This 23.78% is baptized as a group of risk genes, due to its resistance.
Nearly half of the 2,561 antibiotic-resistant genes identified are present simultaneously in diverse habitats, especially those that confer resistance to some of the most widely used antibiotics in use today.
Representatively, almost half of the 2,561 GRAs identified are present simultaneously in diverse habitats, especially genes that confer resistance to some of today's most widely used antibiotics. However, the global distribution of antibiotic resistant genes, based mainly on their number and abundance, is relevant but insufficient to assess the health risk they pose.
One of the objectives of the research is to quantify and standardise the threats of antibiotic resistance worldwide, with the aim of improving decision-making in public health policy. The expert group says that monitoring the risk posed by antibiotic-resistant genes should be a priority for global cooperation.
23.78% of antibiotic-resistant genes confer multi-drug resistance and, as a consequence, make clinical treatments increasingly sophisticated.
The study has been driven by an international team including the CREAF and CSIC researcher Josep Peñuelas and is led by researcher Haifeng Qian from the Zhejiang University of Technology (China). "Antibiotic resistance is further evidence of the drastic change in the human microbiota worldwide", according to Josep Peñuelas, whose scientific dedication continues to explore the 'One health' concept, among many others. The work published in Nature Communications has taken into account the accessibility of GRAs in humans and animals, their mobility, pathogenicity and clinical availability for testing, in a metagenomic analysis of 4,572 samples from 6 different habitats.
How a resistant gene is spread
Human activity is the main disseminator of antibiotic-resistant genes, due to the intensive use of antibiotics to fight bacterial infections. Today, however, resistance itself is not caused by humans, as these genes existed before the antibiotic era and were detected in human faeces in the Palaeolithic and permafrost.
The results obtained imply that human activity is the main disseminator of antibiotic resistant genes, due to the frequent and intensive use of antibiotics to fight bacterial infections, both in medicine and livestock. However, resistance itself is not caused by current human activity, as GRAs existed before the antibiotic era and were already detected in human faeces in the Palaeolithic and permafrost. The current large increase in GRA is attributed to its transmission by viruses (transduction), free DNA (transformation), bacterial linkages (conjugation) and plasmids (autonomously replicating DNA molecules).
To understand how human activity is affecting antibiotic resistance today, the study examined the abundance and composition of GRAs in various habitats on a global scale. Of all the genes identified,the ones found in the greatest number are of human origin, especially those of the digestive system and human skin. These genes reside in the human body, largely located in our healthy microbiota. This presence does not cause health problems in healthy people, but becomes an alarm signal in case of illness.
The samples of genetic material analysed in the research come from a variety of habitats (human, animal, air, aquatic, terrestrial, etc.) including natural, engineered (habitats that manage water that has been in contact with humans, such as water treatment plants) and clinical habitats. Sub-habitats such as buildings and suburban subways, among other infrastructures, are also included in the study.
A global map of the marine habitat
Thanks to machine learning, the presence of these genes in the world's oceans has been examined. This novel technique has produced a global map of the risks posed to human health by the presence of antibiotic resistant genes in global marine habitats. This analysis is estimated to be more than 75% accurate. The study does not determine the origin of these genes in marine waters, but it is a reflection of human and animal activity on land. Thus, the oceans can be said to become sentinels of terrestrial activity linked to antibiotic-resistant genes.
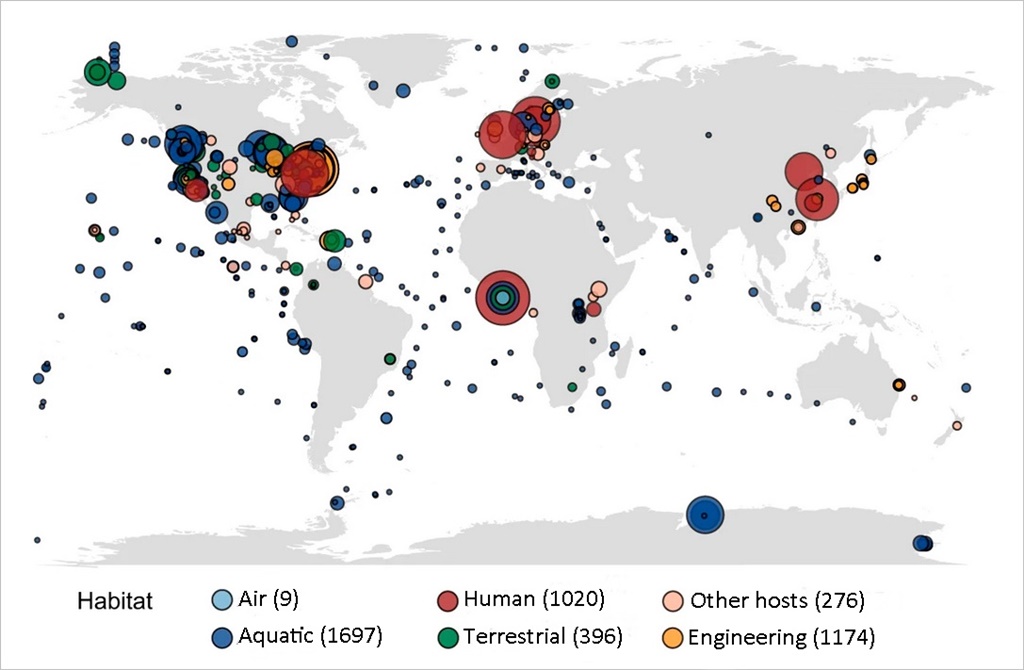
The mapping shows that marine areas near Brazil and Africa have a higher risk of antibiotic-resistant genes. And the comparison between oceans indicates that the threats of resistance to antibiotics in the Pacific and the Atlantic are greater than in the others.
However, it has not been possible to replicate this map for terrestrial habitats, due to the uneven distribution of the samples and the lack of metadata on the original properties of the soil. Soil and deep soil samples with which humans have been in contact were also evaluated, and horizontal transfer of antibiotic resistant genes between humans, animals and freshwater has been detected. The research demonstrates the horizontal transfer of antibiotic-resistant genes via bacteria and DNA between humans, livestock, freshwater and soils with which they come into contact. This round-tripping of antibiotic-resistant genes between soil, water, animals and people around the world already existed, but is intensifying.
Reference:
Assessment of global health risk of antibiotic resistance genes. Zhenyan Zhang, Qi Zhang, Tingzhang Wang, Nuohan Xu, Tao Lu, Wenjie Hong, Josep Penuelas, Michael Gillings, Meixia Wang, Wenwen Gao & Haifeng Qian. Nature Communications, 2022.







